Research - Proteomics
Electrospray–assisted Laser Desorption Ionization (ELDI)Electrowetting–on–dielectric (EWOD)
Human Salivary Proteome
Virtual 2D Gels
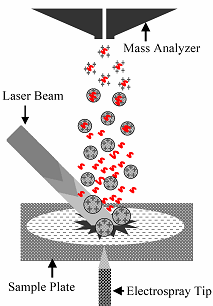
Electrospray–assisted Laser Desorption Ionization (ELDI)
Summary
ELDI is a new, soft ionization method which combines features of both ESI and MALDI. In ELDI, neutral protein molecules are desorbed from solid samples by a nitrogen laser pulse and post-ionized through fusion with charged solvent droplets generated by ESI. The advantages of ELDI include: (1) it works under ambient conditions, (2) laser desorption has a higher power for desorption compared to DESI (Desorption ESI), (3) matrices are unnecessary in ELDI, (4) ESI-like ions give more accurate molecular weight and reduce the mass requirement of the mass analyzer for large protein analysis.
Electrowetting–on–dielectric (EWOD)
Summary
Microfluidics is a technology which can integrate multiple functions onto a single device. Applications of microfluidics include separations, reactions, immunoassays, enzymatic digests, and cell transport and analysis. The capacity for integrating multiple laboratory processes makes microfluidics an attractive technology for proteomics applications.
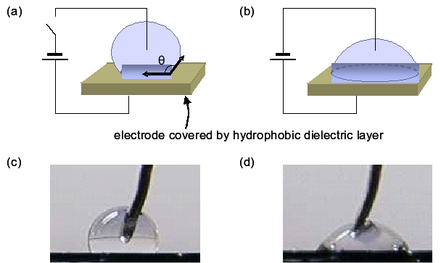
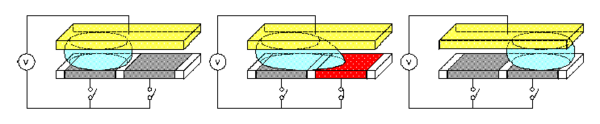
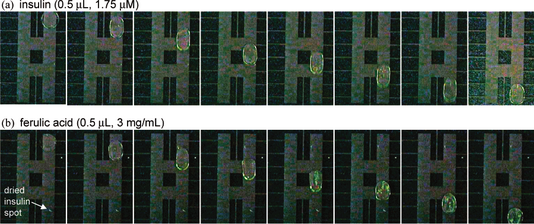
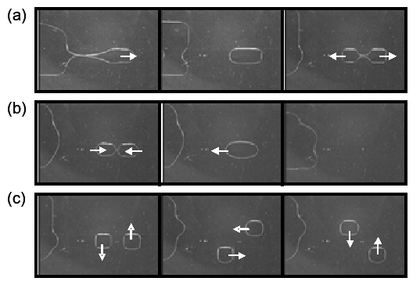
EWOD is a new microfluidic technique which transports fluid as discrete droplets on a surface. The movement of droplets on the EWOD surface is based on the buildup of charge on the dielectric material, which makes the surface more hydrophilic and decreases the contact angle of the droplet, θ. EWOD chip can serve as a media for protein purification and is comparable to MALDI-TOF. In collaboration with Professor Robin Garrell, we hope to develop EWOD into a useful technique for high-throughput proteomic studies.
Publications
Wheeler AR, Moon H, Bird CA, Ogorzalek Loo RR, Kim CJ, Loo JA, and Garrell RL. Digital Microfluidics with In-line Sample Purification for Proteomics Analyses with MALDI-MS.
Anal Chem 2005; 77: 534-540.
Wheeler AR, Moon H, Bird CA, Ogorzalek Loo RR, Kim CJ, Loo JA, and Garrell RL. Electrowetting-Based Microfluidics for Analysis of Peptides and Proteins by Matrix-Assisted Laser Desorption/Ionization Mass Spectrometry.
Anal Chem 2004; 76: 4833-4838.
Human Salivary Proteome
Summary
The Human Salivary Proteome Project is an effort to generate a catalog of proteins and their posttranslational modifications in the secretions from the three major salivary glands namely parotid, submandibular and sublingual. The tools currently being employed to map the salivary proteome include:
- Shotgun Proteomics. In the shotgun approach salivary proteins are first prefractionated either by their molecular weight using a MW cutoff filter or by their pI by in-solution Zoom IEF fractionation. The proteins in each fraction are then digested with trypsin. The digests are further analyzed and proteins are identified by 1D and 2D-LC mass spectrometry.
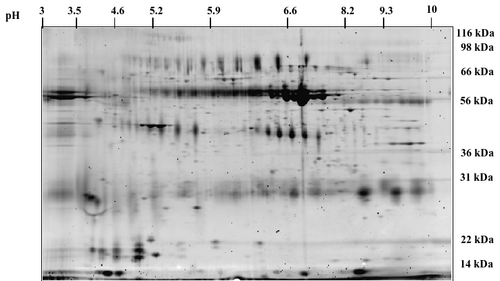
- Two-dimensional gel electrophoresis-mass spectrometry (2-DE-MS). Proteins from the saliva are separated on a 2D-gel. The gel is stained with total protein stain like Sypro Ruby. The gel spots are then excised, proteins therein are digested in-gel and identified by mass spectrometry.
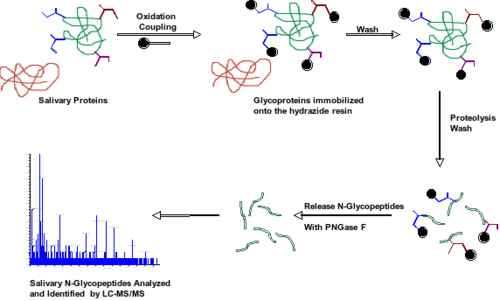
We also aim to identify posttranslational modifications on salivary proteins like glycosylation and phosphorylation. The technique currently being employed to identify N-glycosylated proteins is the hydrazide coupling and release method. In this approach, glycoproteins are coupled onto a hydrazide resin, the proteins are then digested and formerly N-glycosylated peptides are selectively released with the enzyme PNGase F and analyzed by LC-MS/MS. Employing this method we have identified more than 40 N-linked salivary glycoproteins.
Publications
Hu S, Li Y, Wang J, Xie Y, Tjon K, Wolinsky L, Ogorzalek Loo RR, Loo JA, and Wong DT. Human Saliva Proteome and Transcriptome.
J Dent Res 2006; 85: 1129-1133.
Hu S, Loo JA, and Wong DT. Human body fluid proteome analysis.
Proteomics 2006; 6: 6326-6353.
Ramachandran P, Boontheung P, Xie Y, Sondej M, Wong DT, and Loo JA. Identification of N-Linked Glycoproteins in Human Saliva by Glycoprotein Capture and Mass Spectrometry.
J Proteome Res, 2006; 5: 1493-1503.
Hu S, Xie Y, Ramachandran P, Ogorzalek Loo RR, Li Y, Loo JA, and Wong DT. Large-Scale Identification of Human Oral Fluid Proteins by Liquid Chromatography-Mass Spectrometry.
Proteomics 2005; 5: 1714-1728.
Virtual 2D Gel Electrophoresis
Summary
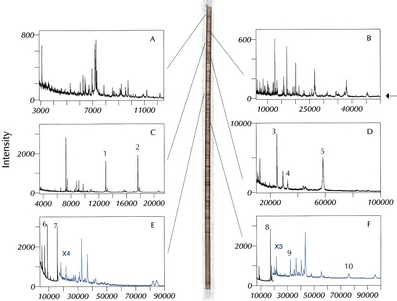
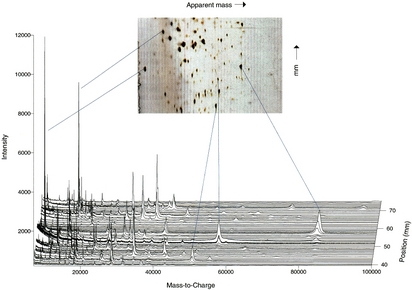
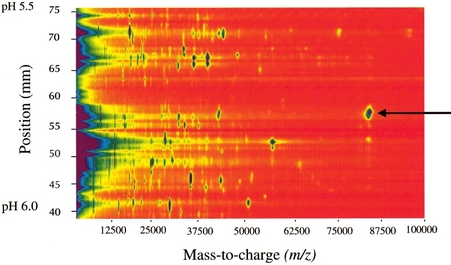
In the virtual 2D gel
approach, mass spectrometry is
substituted for SDS-PAGE size-based separation. Use of mass
spectrometry for the second dimension provides advantages in mass
resolution, mass accuracy, and sensitivity over classical 2D gels.
Post-translational modifications can be readily detected by the
mass-resolved heterogeneity of component hydrocarbon chains. Direct
comparison to classical 2D gels provides a link between 2D gel spots
and accurate, higher resolution molecular weight measurements.
Publications
Ogorzalek Loo RR, Hayes R, Yang Y, Hung F, Ramachandran P, Kim N, Gunsalus R, and Loo JA. Top-Down, Bottom-Up, and Side-to-Side Proteomics with Virtual 2-D Gels.
Int J Mass Spectrom 2005; 240: 317-325.
Ogorzalek Loo RR, Yam L, Loo JA, and Schumaker VN. Virtual 2-D Gel Electrophoresis of High Density Lipoproteins.
Electrophoresis 2004; 25: 2384-2391.
Ogorzalek Loo RR, Cavalcoli JD, VanBogelen RA, Mitchell C, Loo JA, Moldover B, and Andrews PC. Virtual 2-D Gel Electrophoresis: Visualization and Analysis of the E. coli Proteome by Mass Spectrometry.
Anal Chem 2001; 73: 4063-4070.
Ogorzalek Loo RR, Direct Molecular Weight Determination of Gel Separated Proteins
, in Encyclopedia of Mass Spectrometry, M. Gross and R. Caprioli, Eds., Elsevier 2004; 36-47.
Loo JA, Brown J, Critchley G, Mitchell C, Andrews PC, and Ogorzalek Loo RR, High Sensitivity Mass Spectrometric Methods for Obtaining Intact Molecular Weights from Gel-Separated Proteins
, Electrophoresis 1999; 20: 743-748.
Ogorzalek Loo RR, Mitchell C, Andrews PC, VanBogelen RA, and Loo JA, MALDI Visualization of the E. Coli Proteome
, Proceedings of the 1st Joint Services Workshop on Biological Mass Spectrometry 1998; 91-96.
Ogorzalek Loo RR, Loo JA, and Andrews PC, Obtaining Molecular Weights of Proteins and Their Cleavage Products by Directly Combining Gel Electrophoresis with Mass Spectrometry
, in Methods in Molecular Biology: 2-D Protein Gel Electrophoresis Protocols, A. Link, Ed; Humana Press: Totowa, NJ, pp. 473-485 (1998).
Ogorzalek Loo RR, Loo JA, Andrews PC, Gels in Vacuo? A Minimalist Approach for Combining Mass Spectrometry and Polyacrylamide Gel Electrophoresis
, in Mass Spectrometry of Biological Materials, 2nd Ed., B. S. Larsen and C. N. McEwen, Eds.; Marcel Dekker: New York, pp. 325-343 (1998).
Ogorzalek Loo RR, Mitchell C, Stevenson TI, Loo JA, and Andrews PC, Diffusive Transfer to Membranes as an Effective Interface Between Gel Electrophoresis and Mass Spectrometry
, International Journal Mass Spectrometry and Ion Proc. 169/170, 273-290 (1997).
Ogorzalek Loo RR, Mitchell C, Stevenson TI, Martin SA, Hines W, Juhasz P, Patterson D, Peltier J, Loo JA, and Andrews PC, Sensitivity and Mass Accuracy for Proteins Analyzed Directly from Polyacrylamide Gels: Implications for Proteome Mapping
, Electrophoresis1997; 18: 382-390.
Ogorzalek Loo RR, Stevenson TI, Mitchell C, Loo JA, and Andrews PC, Mass Spectrometry of Proteins Directly from Polyacrylamide Gels
, Anal Chem 1996; 68: 1910-1917.
Ogorzalek Loo RR, Mitchell C, Stevenson TI, Loo JA, and Andrews PC, Interfacing Polyacrylamide Gel Electrophoresis with Mass Spectrometry
, in Techniques in Protein Chemistry VII, D. Marshak, Ed; Academic Press: San Diego, CA, pp. 305-313 (1996).
